m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover

Relevance of N6-methyladenosine regulators for transcriptome: Implications for development and the cardiovascular system - ScienceDirect

ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells | PNAS
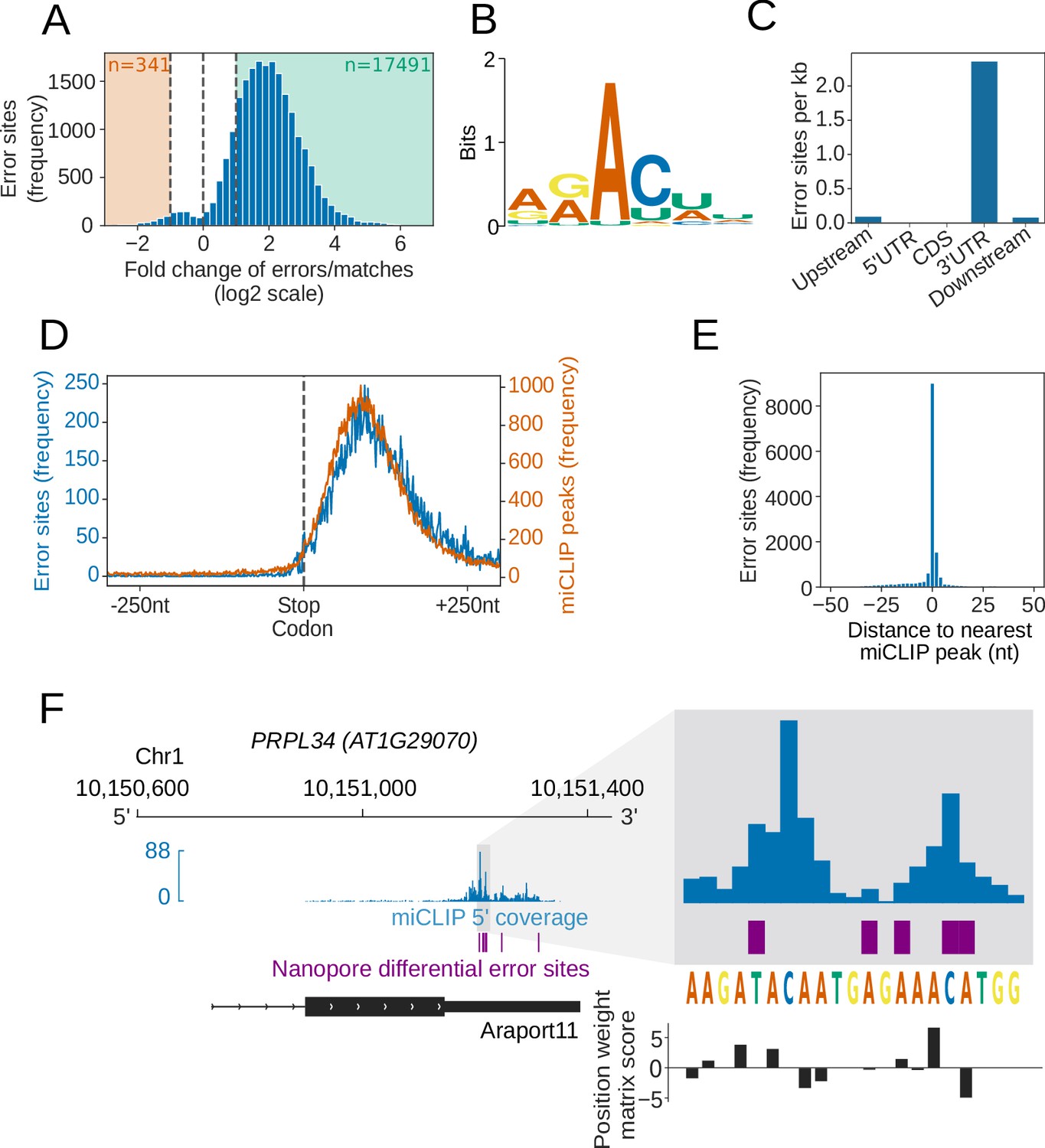
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife

Single-nucleotide resolution achieved by m 6 A-CLIP/immunoprecipitation... | Download Scientific Diagram

N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. – 武汉生命之美科技有限公司

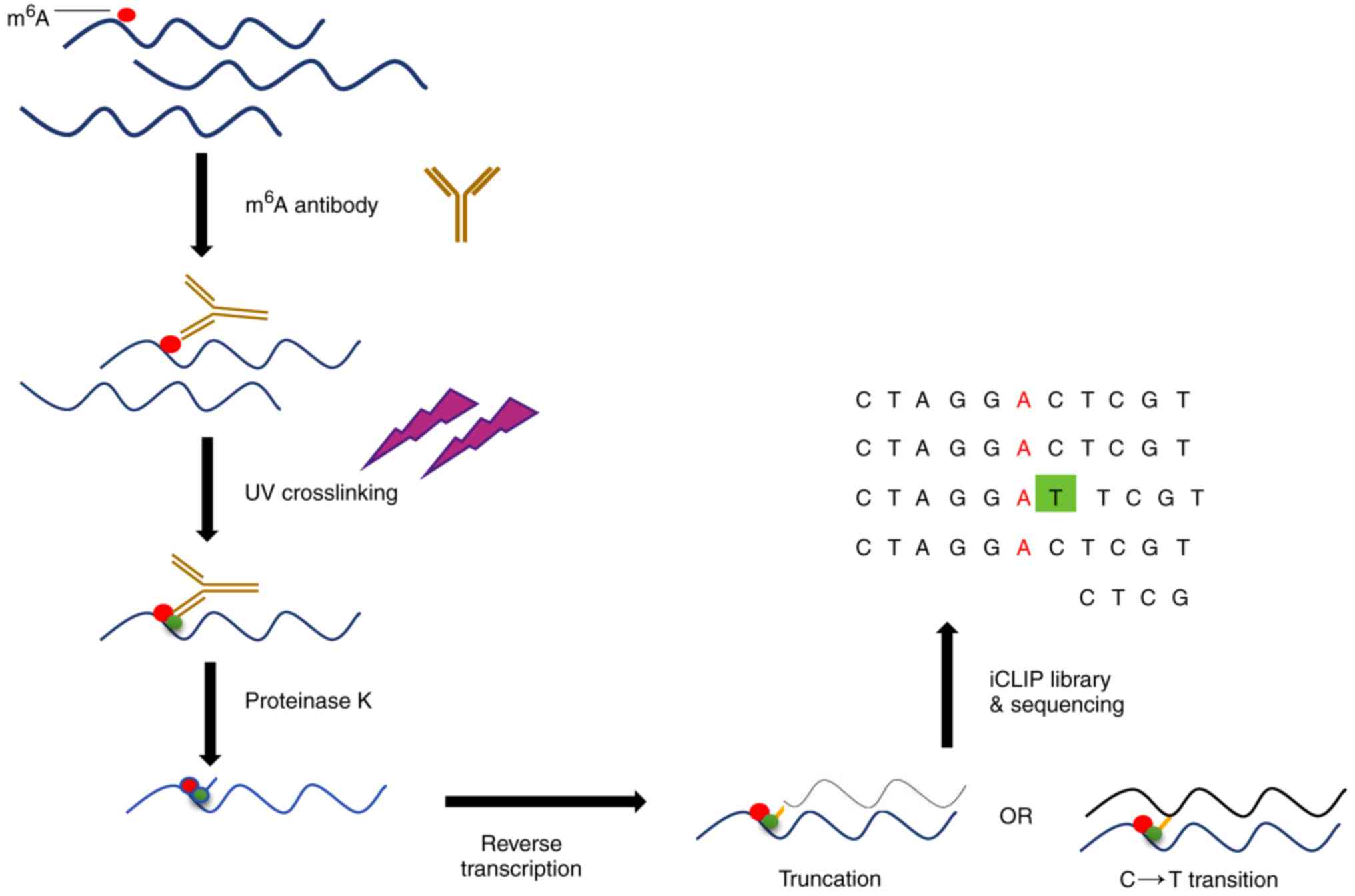
Deep and accurate detection of m6A RNA modifications using miCLIP2 and m6Aboost machine learning | bioRxiv

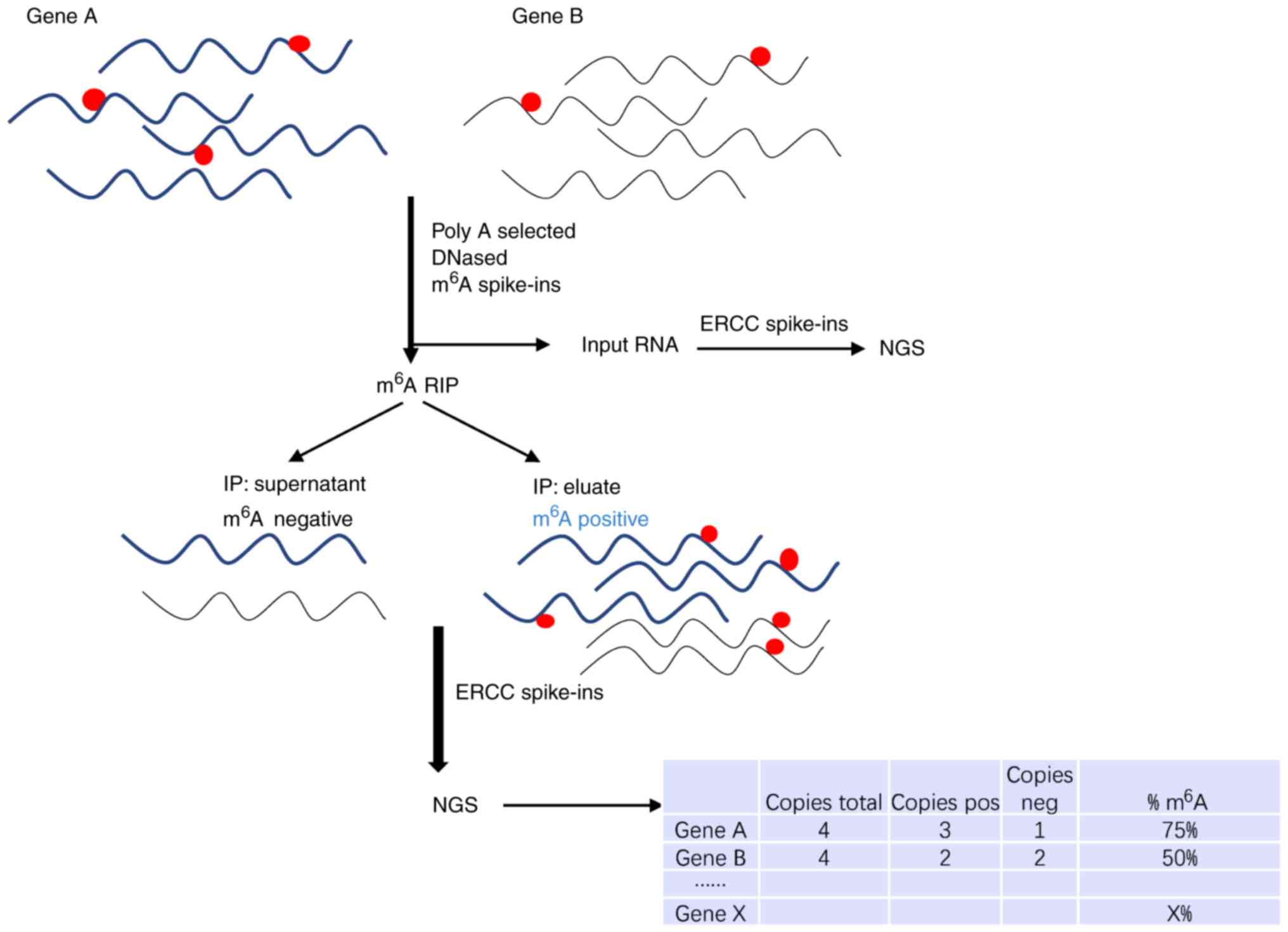
Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing | Nature Protocols

Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine | Nature Chemical Biology

Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation - ScienceDirect

m6A deposition is regulated by PRMT1‐mediated arginine methylation of METTL14 in its disordered C‐terminal region | The EMBO Journal